Snakemake: A Hidden Gem for Sustainable Data Science from the Field of Bioinformatics
Problem analysis
Snakemake is an open source workflow management system inspired by the GNU Make [^gnu-make] build automation tool. Snakemake aims to facilitate sustainable data analysis by supporting reproducible, adaptable, and transparent data research. This is done by using workflows, which are data analysis pipelines. The core of a Snakemake workflow consists of a Snakefile that defines all the steps of a workflow as rules. These rules determine how output files are created from input files, while Snakemake automatically resolves dependencies between the rules. For example, rules can be used to edit input data or generate figures. By using Snakemake developers can create reproducible and scalable data analysis pipelines in Python.
The rolling paper accompanying Snakemake identifies five different types of workflow management systems that can be used for automated data analysis 1. They differ in the level of knowledge required in order to operate them. For instance, workflow management systems like Galaxy and KNIME employ a graphical interface in order to be accessible for a larger part of the scientific community, including those that do not have a background in computer science or programming. Other systems, like CWL and WDL, opt to maximize scalability, by creating a declarative language which sets a standard for data analysis. The implementation of that analysis is then left to other programs.
Snakemake opts to maximize usability instead. Being written as an extension to the Python language, Snakemake has access to the full power of the Python programming language, while still being easily readable by means of the Snakemake constructs/syntax, removing the need for any boilerplate code for workflow management.
In this essay we offer an analysis of the quality attributes Snakemake strives to meet and why the widespread adoption, based on these attributes, seems to be lacking. It is part of a series of four essays on the architecture of Snakemake.
Beyond reproducibility
As mentioned before, Snakemake aims to provide the researcher with the means to analyze data in a reproducible, transparent and adaptable fashion. Thus, the authors of the Snakemake paper look further than reproducibility, stating that “reproducibility alone is by no means sufficient to deliver an analysis that is of lasting impact (i.e., sustainable) for the field.” 2. Adaptability and transparency are both important to ensure sustainable data research, according to the authors. Figure 1 shows their hierarchy of values for enabling these three aspects. Below we will explain these values, as well as their relation in more detail.
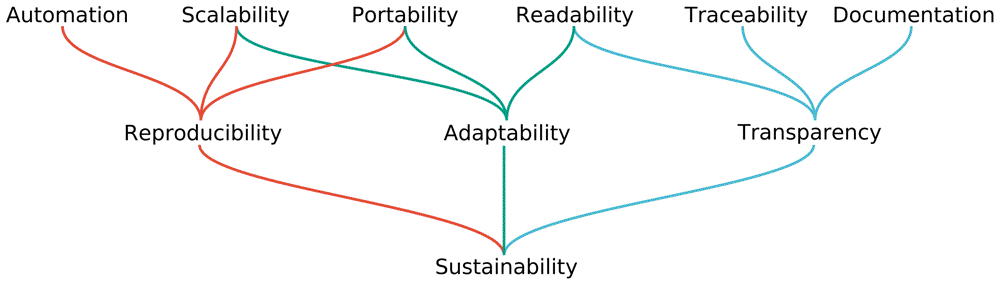
First of all, the obvious: Reproducibility. In order to make data analysis reproducible, the processes guiding reproducibility have to be automated in a deterministic, consistent fashion. Furthermore, the data analysis needs to be scalable to multiple computational platforms. This ensures the data analysis is reproducible amongst a greater part of the scientific community. This value of scalability is related to portability: The software should be easily deployable without prior setup. This also ensures the data analysis is reproducible by a greater part of the scientific community, as scientists less versed in informatics are then also able to reproduce the data analysis.
The second pillar is adaptability; how well you can reuse some parts of the analysis for other projects or on different hardware. Since different projects require different tools, the workflow is usually not copied one to one, but instead adapted for different purposes. In order to allow for adaptability, the workflow also needs to be scalable and portable to different systems, like for the reproducibility pillar. This makes it easy to adapt the workflow to a different project on a different system. As part of portability, parts of the workflow require specific tools to be installed and configured. This requires these parts of the workflow to be executed in a containerized manner. A third important value for adaptability is creating readable code. After all, if it’s very difficult for a scientist to understand what a piece of code does, it makes it difficult to adapt said code to a different project.
This value of readability feeds into the third pillar which Snakemake identifies as important for sustainable data analysis: Transparency. If code is unreadable, it does not allow for other scientists to accurately gauge if the data analysis is performed as it is stated to be. Good documentation also allows for greater transparency. Lastly, it is important for code, parameters and components to be traceable through all involved steps.
Why aren’t more researchers using Snakemake?
The documentation describes Snakemake as “a tool to create reproducible and scalable data analyses” 2, without pointing to a specific (scientific) field as its intended user base. Indeed, it seems that Snakemake offers a lot of value to anyone doing serious data analysis. However, looking at the data in Figure 2, we notice Snakemake is used by bioinformaticians mostly. Over 50% of all citations are from bioinformatics papers. Meanwhile, citations from other computer science papers make up only 14% of total citations. Why is this? Why is adoption in other fields that require rigorous data analysis lacking compared to bioinformatics?
While we postulate our own causes here, we also reached out to the founder and main developer of Snakemake, as well as author of the paper 3, Johannes Köster. He was able to give us some quick comments, and we will mention some of his first thoughts on the question here as well.

The startup view
The difference in adoption could simply be a matter of marketing and distribution. Although Snakemake is not a commercial product, it faces similar challenges any startup would, assuming the goal is to expand Snakemake to other fields.
As Köster pointed out, the original paper on Snakemake 1 was published in the bioinformatics domain. Furthermore, the tutorials 2 and examples available on the internet are all about bioinformatics applications.
One could argue that, since Snakemake is a great ‘product’ that offers a lot of value to most computer science researchers, naturally, the adoption of Snakemake in other fields is simply a matter of time. The product may sell itself. However, we think that this way of reasoning is a mistake, one that is especially prevalent among engineers. The system for marketing and distributing the ‘product’ should be viewed as an integral part of the product itself.
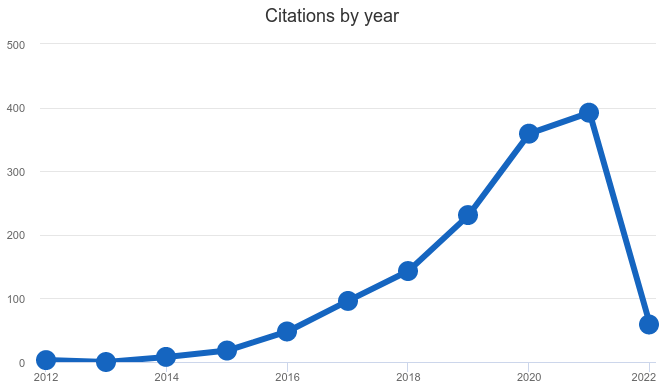
Therefore, in our view, the lack of publicity in other domains than bioinformatics (marketing), and the lack proper tutorials that appeal to non-bioinformatics researchers (distribution), means that it is likely that Snakemake will not automatically be adopted in other fields. We can see in Figure 3 the number of citations per year. In 2021 the number of publications was not in line with the past exponential growth. Whether this hints at the rate of adoption stalling, or is just an outlier, for instance due to some other phenomenon impacting the rate of publications (such as the pandemic) can not be determined here.
The historic view
Other aspects that could explain the difference in adoption are historic in nature. According to Köster, fields like machine learning established their means of managing workflows before open source development became a major factor of consideration. Furthermore, as suggested by Köster, data analysis in bioinformatics has traditionally been heterogeneous (meaning that one requires a lot of different tools to do the job), thereby increasing the need for a generic workflow management system like Snakemake.
Existing machine/deep learning frameworks are more self contained, often relying on a single programming language, reducing the need for a workflow manager. This is illustrated by the common use of Jupyter notebooks in such fields. In other words, perhaps researchers in other fields have acceptable alternatives to Snakemake, and thus more effort is required to convince them of the benefits of using Snakemake (returning again to our previous point of marketing and distribution). We certainly think there are some great advantages of using Snakemake over notebooks. For example, Snakemake tracks its data files for timestamps and can automatically track if a file changes what specific parts must be re-run instead of re-running the complete pipeline.
Lastly, Köster mentioned that bioinformatics research might place an increased emphasis on reproducibility. This might be explained by the often noisy and difficult to interpret datasets that stand at the basis of bioinformatics research. However, this remains speculation on our part.
The developer’s view
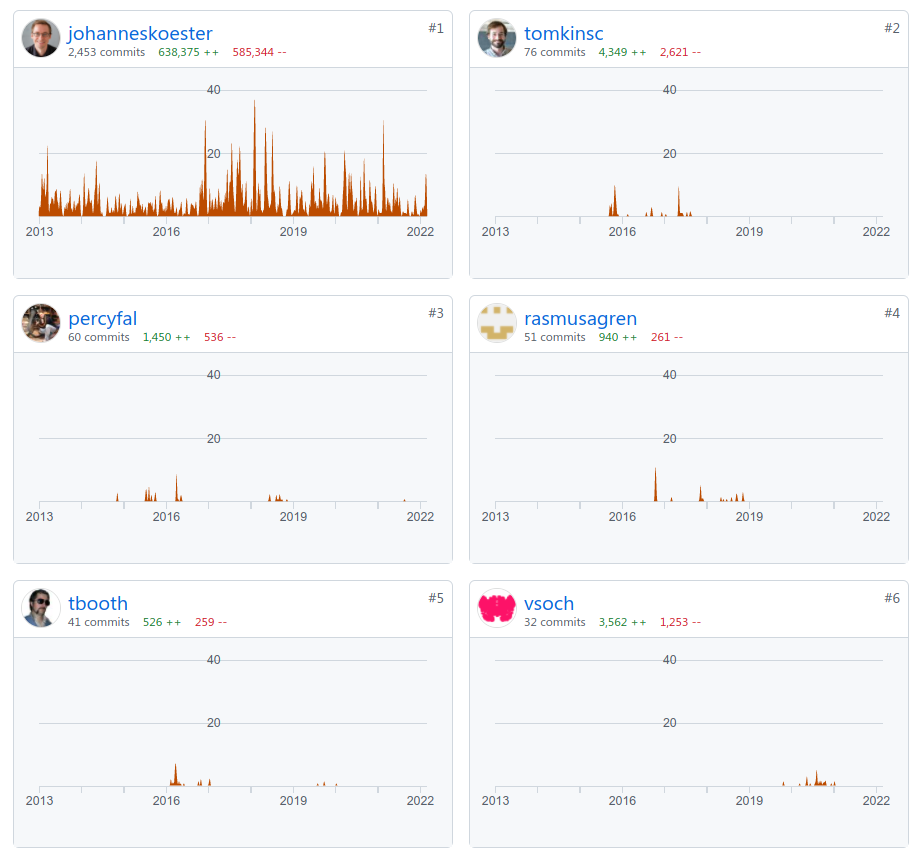
The last factor that could contribute to the difference in adoption is about development. Köster is by far the biggest contributor to the Snakemake project, as can be seen in Figure 4. Since his main interests are in the field of bioinformatics 4, we think it is logical for development, documentation and tutorials to be focused on bioinformatics. For example, the tutorials use bioinformatics applications as examples, and there are sections of the documentation focused solely on bioinformatics applications, such as executing workflows via Google Cloud Life Sciences 5.
However, for potential users outside the bioinformatics field, seeing that development is focused on the field of bioinformatics might discourage them from trying out Snakemake. Moreover, even if they are convinced of Snakemake’s usefulness, a user considering contributing a new feature might not feel as incentivized to do so if it is not meant for the field of bioinformatics.
The future of Snakemake
As of the moment of writing, automatic translation of CWL into Snakemake is planned 3. This feature would also lead to higher adaptability. Furthermore, version 7 of Snakemake was recently released adding scalability features.
Snakemake should do well to refine their target audience going forward in our opinion. When focusing solely on the field of bioinformatics, greater domain-specific functionality can be achieved. Another path Snakemake can take is to try to expand into other fields. In this case, usability in other fields can be maintained, for instance through the use of plugins. This allows Snakemake to be usable in other fields, while not distracting the developers and main user base with feature overload. Our next essays further investigate this possibility.
References
-
J. Koster and S. Rahmann, “Snakemake–a scalable bioinformatics workflow engine,” Bioinformatics, vol. 28, no. 19. Oxford University Press (OUP), pp. 2520–2522, Aug. 20, 2012. doi: 10.1093/bioinformatics/bts480. ↩︎
-
J. Köster, “Snakemake — Snakemake 7.1.0 documentation”, Snakemake.readthedocs.io, 2022. [Online]. Available: https://snakemake.readthedocs.io/en/stable/. [Accessed: 07-Mar-2022]. ↩︎
-
F. Mölder et al., “Sustainable data analysis with Snakemake” F1000Research, vol. 10. F1000 Research Ltd, p. 33, Apr. 19, 2021. doi: 10.12688/f1000research.29032.2. ↩︎
-
J. Köster, “Johannes Köster’s Homepage”, Johanneskoester.bitbucket.io, 2022. [Online]. Available: https://johanneskoester.bitbucket.io/. [Accessed: 07-Mar-2022]. ↩︎
-
J. Köster, “Cloud execution — Snakemake 7.1.0 documentation”, Snakemake.readthedocs.io, 2022. [Online]. Available: https://snakemake.readthedocs.io/en/stable/executing/cloud.html#executing-a-snakemake-workflow-via-google-cloud-life-sciences. [Accessed: 07-Mar-2022]. ↩︎