Introduction
Snakemake is an open source workflow management system inspired by the GNU Make 1 build automation tool. Snakemake aims to facilitate sustainable data analysis by supporting reproducible, adaptable, and transparent data research. The core of a Snakemake workflow consists of a Snakefile that defines all the steps of a workflow as rules. These rules determine how output files are created from input files, while Snakemake automatically resolves dependencies between the rules. For example, the rules can be used to edit input data or generate figures. In this essay we provide an analysis of the architectural design of Snakemake.
This essay is based on an interview we conducted with the code owner of Snakemake, Johannes Köster, as well as our own research into the repository of Snakemake. It is part of a series of four essays on the software architecture of Snakemake.

Dr. rer. nat. Johannes Köster, code owner of Snakemake
History of Snakemake
The main developer behind Snakemake, Johannes Köster, started working on Snakemake in 2012. At the time there were not many options for coding computational workflows. People mainly resorted to the general purpose tool GNU make. GNU make is not intended for data processing but as a build tool for generating executables. It has a steep learning curve due to its cryptic syntax. Thus, Köster created Snakemake, providing a human readable make tool for programming computation workflows.
Behind the scenes
This section provides an overview for all the components, by going through the processes that happen when executing Snakemake. Only the most important and interesting components are addressed for brevity, and they appear in the order they are executed.
Configuration
There are many ways to configure the Snakemake execution. First, one can provide configuration via the command line interface (CLI) with command line arguments. The number of parameters provided over the CLI can get very large 2. That’s why there are additional ways to provide configuration.
You can provide different kinds of configuration with the use of configuration files. There are profile files meant to store cluster configurations. There is pepfile support for describing different sample meta data. PEP here stands for Portable Encapsulated Projects 3 and should not be confused with Python Enhancement Proposals. The pepfile should only be used to describe information about the data, and should be independent from other configuration options. There is also a generic configuration file that defines information specific for this workflow only.
Lastly, there is a way to provide configuration information via system environment variables. Currently, these are only recommended for passing secret information only, as this method is not transparent in use and not very robust, as it has to manually specified upon execution.
The Snakemake parser
A central component for Snakemake is the parsing of the Snakefiles. This is one component of the codebase that has seen the least amount of change over the years according to Köster. As this is a core functionality of Snakemake, we will dive deeper into the parsing and transpilation components in this section.
The Snakefile defines rules with which to create output files from input files. It is the central spot to define your Snakemake workflow. The Snakemake parser takes the Snakefile and transpiles it to pure Python. This transpiled Python file is then used by Snakemake to infer the directed acyclic graph (DAG) of jobs.
Since the Snakemake language is an extension of Python, an interesting design decision was made to only transpile the Snakemake-specific code and leave the existing python code inside the Snakefile untouched. In principle, this means that the Snakemake parser is indifferent to Python versions, so long as the Python version supports decorators (they are used extensively in the transpiled code).
The current approach for parsing the Snakefile is quite interesting, although we think a different approach would net some important benefits, without compromising on current capabilities. Let us first take a look at the current approach.
Current approach
Currently, the parser takes an ad-hoc approach. It processes the Snakefile line by line, transpiling detected Snakemake constructs on the fly. No abstract syntax tree (AST) is created as an intermediate semantic representation of the Snakefile’s contents. Nevertheless, the parsing components have a clear structure, and in our eyes the current solution is already quite elegant, using a concept dubbed by Köster as the “automaton”.
An automaton is responsible for transpiling a specific type of construct in the Snakefile. For example, there is an automaton for transpiling a ‘rule’ construct (see figure 1). It will relinquish transpiling duties to its parent automaton once it reaches the end of its designated construct. These ends-of-constructs are easily detected from the identation. If the automaton encounters a nested Snakemake construct, such as the input/output keywords within a rule, it will spin up a child automaton to take over transpiling until the nested construct is transpiled. The root automaton is the ‘Python’ automaton, which leaves pure python code untouched and hands over transpilation to its child automata when it encounters a Snakemake construct.
The Snakemake rules are transpiled into Python functions. Since no AST is built, there needs to be another way to interpret the Snakemake constructs and infer jobs and the DAG. This is accomplished by decorating the transpiled rules functions with decorators from the workflow component. These decorators capture important information such as input and output to a rule, but they also serve to register/store the various rules in a central spot (an instance of the Workflow class).
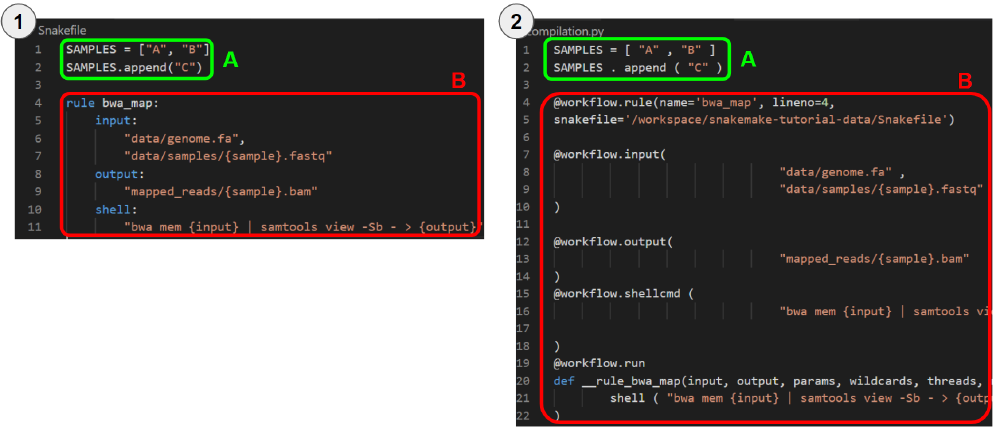
Figure 1. In figure 1.1, we show an example of a simple Snakefile with a single rule. In figure 1.2, we show the transpilation result.
To conclude, the parser in its current form is not only a parser, but also a transpiler at the same time. In figure 2, we give a visual overview of the components that are involved and their relations. While clearly structured, we see some disadvantages to the current approach. Firstly, the code for parsing, giving parsing errors and transpilation are in the same place, reducing readability. Secondly, the workflow component depends on the fact that the parser/transpiler uses the workflow function decorators correctly. This makes it difficult to introduce alternative parsers that would make other workflow management languages usable with Snakemake.
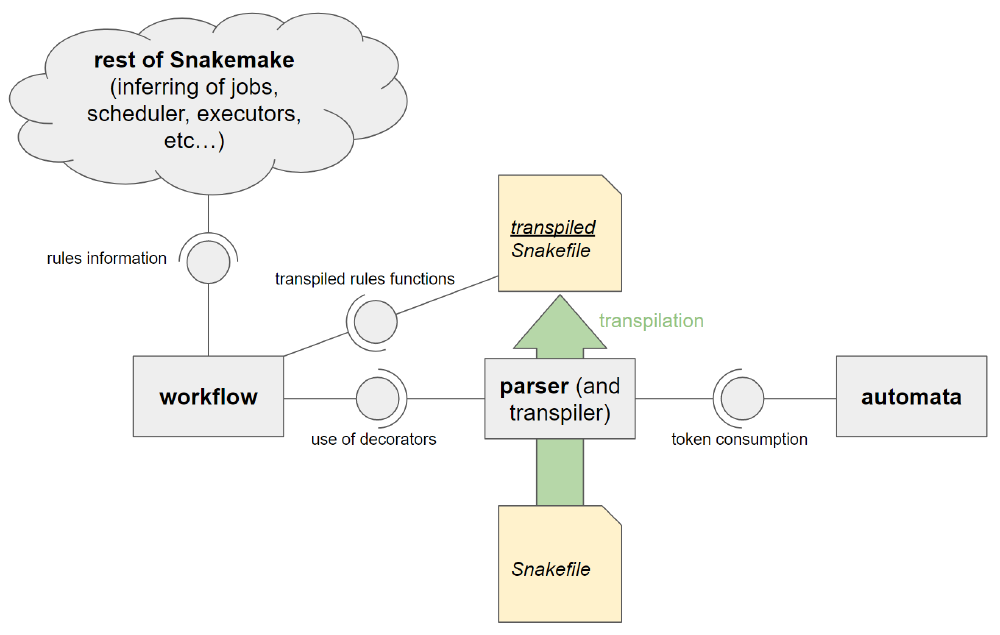
Figure 2. The current component level view of the parsing and transpilation system.
Suggested approach
We think an alternative approach could have some benefits. Instead of an ad-hoc approach, we propose to build an AST representation of the Snakefile. To retain the current parser’s indifference to Python versions, we would define one node type of the AST to be “Python code that should be left untouched”. A separate transpiler component would use the AST to produce the transpiled Snakefile. The workflow component would use the AST and transpiled Snakefile to gather the rule info just as before.
We believe the main benefits of this approach are 1) the parser and transpiler are decoupled, 2) it elegantly facilitates semantic code analysis of Snakefiles (for linting, detecting code smells, etc.), 3) as Köster mentioned during the interview, he would like to support testing of Snakemake workflows with a way to show test coverage, but currently this would be difficult to accomplish. Having an AST is a clear starting point for being better to run test on the rules, as the relation between Snakefile code and pure python code (the code that is actually executed) is much clearer and easier to keep track of.
The decoupling of the parser and the transpiler allow for better testing, and thus addresses the core values of adaptability and transparency, as better testing makes it more transparent to others what a Snakemake workflow does, and also, properly tested Snakefile rules can be more easily adapted to other projects.

Figure 3. Our proposed component level view of the parsing and transpilation system.
Generating a plan from rules and already present files
When all the rules are parsed, the plan for execution must be defined. Each rule defines a certain output and input. The combined inputs for a given rule form the dependents for that rule. All these rules then form a DAG of jobs. This DAG can then be used to see what jobs should wait for other jobs to finish or what jobs should be scheduled first.
The complete plan for execution does not have to be run. Some of the required files might already be present from earlier runs. One of the features of Snakemake is that it keeps track of what files are already created and when those files were created. This way Snakemake only has to run certain jobs if the output files are not present or if the input files for that job are newer than its output files.
The way of checking timestamps can be somewhat unreliable especially if those files live on remote file systems. An interesting feature to include in the future is instead to save a hash of each file to see if it has been renewed or not. This has an additional benefit that if a file is renewed but the contents are not changed, the jobs that depend on that file are not triggered. The downside is that hashing can be very expensive for very large files.
Not all files have to be present on the same filesystem. Files can also be stored on remote locations. There are several connectors present in the main codebase that allow Snakemake to interact with files on all different kinds of technologies such as Dropbox4, AWS S35, WebDav6, or FTP7.
Executing the plan
From the DAG can be extracted what jobs or processes still have to be run to create the desired results. The scheduler then tries to run jobs that are first in the topological ordering (created by the DAG), but also takes into account the resources each job needs and passes this information on to the connected High Performance Cluster (HPC). For this it uses the required resources as defined for each rule.
The scheduler makes use of Mixed Integer Linear Programming (MILP) to create the best scheduling given the constraints. This is a relatively new feature of Snakemake. Before, it used a greedy approach. The most important benefit of this new method is that it can now also add the lifetime of temporary files to the constraints, thus reducing the amount of time a temporary file is present during execution8.
Each job can define a specific isolated software environment in which the job is run. It can specify this using a Conda9 environment file, which can then install the required software dependencies. Alternatively, you can also specify the isolated software environments via containerization technologies such as Docker10 or Singularity11. In the future we might also see support for for example Podman12 or LPMX13. Additionally, all Conda dependent jobs can also run in their own deployed container.
Alternatively there are more ways to define how a job should be run. There are for example wrappers that predefine common rules from the field. There are also jobs that run from a script in the following languages: Python, R, Rust and Julia[^Snakemake-docs].

Figure 3. The current component overview of Snakemake.
Future direction
What we find in the Snakemake codebase is a complex and rich set of components for workflow management. It is mostly well separated with well-defined APIs between the different components. However, we believe some of the components can be split off from the codebase and be maintained separately. For example, the executors, the remote files, or perhaps even the containerization could be separated. This would reduce the burden for the main code maintainer, Köster, to maintain all these components. These components can then be installed separately as a plugin. It also encourages people to write their own plugins, or replace certain buggy plugins with their own versions. Especially, people or companies introducing new technologies in the HPC field now get the opportunity to write their own plugins for Snakemake.
References
-
J. Köster, “Snakemake — Snakemake 7.1.0 documentation”, Snakemake.readthedocs.io, 2022. [Online]. Available: https://snakemake.readthedocs.io/en/stable/. [Accessed: 07-Mar-2022]. ↩︎
-
- F. Mölder et al., “Sustainable data analysis with Snakemake” F1000Research, vol. 10. F1000 Research Ltd, p. 33, Apr. 19, 2021. doi: 10.12688/f1000research.29032.2.